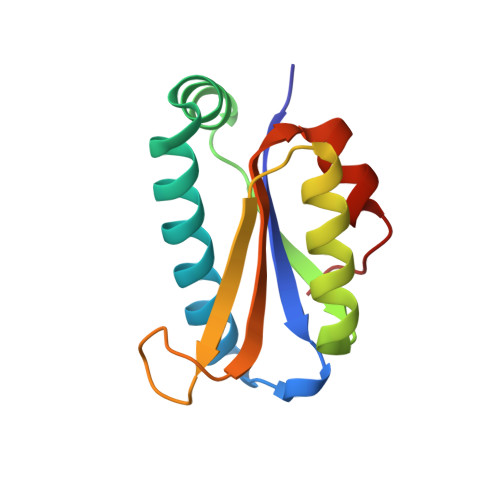
Crystal structure of a human peptidyl-tRNA hydrolase reveals a new fold and suggests basis for a bifunctional activity.
De Pereda, J.M., Waas, W.F., Jan, Y., Ruoslahti, E., Schimmel, P., Pascual, J.(2004) J Biol Chem 279: 8111-8115
- PubMed: 14660562
- DOI: https://doi.org/10.1074/jbc.M311449200
- Primary Citation of Related Structures:
1Q7S - PubMed Abstract:
Peptidyl-tRNA hydrolase (Pth) activity releases tRNA from the premature translation termination product peptidyl-tRNA. Two different enzymes have been reported to encode such activity, Pth present in bacteria and eukaryotes and Pth2 present in archaea and eukaryotes. Here we report the crystallographic structure of the Homo sapiens Pth2 at a 2.0-A resolution as well as its catalytic properties. In contrast to the structure of Escherichia coli Pth, H. sapiens Pth2 has an alpha/beta fold with a four-stranded antiparallel beta-sheet in its core surrounded by two alpha-helices on each side. This arrangement of secondary structure elements generates a fold not previously reported. Its catalytic efficiency is comparable with that reported for the archaeal Sulfolobus solfataricus Pth2 and higher than that of the bacterial E. coli Pth. Several lines of evidence target the active site to two close loops with highly conserved residues. This active site architecture is unrelated to that of E. coli Pth. In addition, intermolecular contacts in the crystal asymmetric unit cell suggest a likely surface for protein-protein interactions related to the Pth2-mediated apoptosis.
Organizational Affiliation:
Program on Cell Adhesion and Signal Transduction, Cancer Center of the Burnham Institute, La Jolla, California 92037, USA.